
Contact
Toxoplasma gondii is an intracellular pathogen that has infected over a billion people worldwide. This parasite sets up a chronic infection in its host that cannot be cured with currently available chemotherapies. Chronically infected humans that become immunocompromised due to HIV/AIDS are at high risk to develop severe toxoplasmosis that can lead to encephalitis and death. Our long term goal is to identify factors in the parasite that are important in the host-pathogen interaction and have the potential to become new therapeutic or vaccine targets. We do this using a variety of approaches.
Toxoplasma is closely related to other pathogens with a significant impact on human health, including the causative agents of malaria and cryptosporidiosis. What distinguishes Toxoplasma from these organisms is its incredible ability to infect a wide array of hosts, ranging from birds to humans. We hypothesize that the genes underlying this unique feature play important roles in human infections.
These unique features of Toxoplasma bring up a number of important questions:
- What evolutionary events produced a parasite species with such a broad host range and how has that impacted its global success?
- Why do different Toxoplasma strains have different levels of virulence and/or compatibility in different hosts?
- What is the link between host range expansion and pathogenesis?
Fig 1: Toxoplasma secretion is mediated by unique secretory organelles such as the rhoptries and dense granules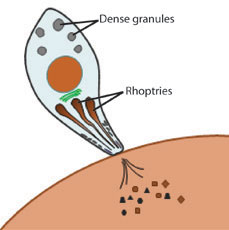
Fig 2: After invasion Toxoplasma takes up residence within the parasitophorous vacuole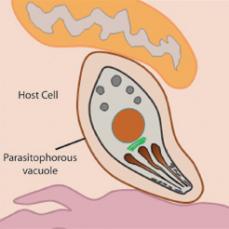
We can now begin to directly address these questions based the recent identification of parasite effector proteins that determine virulence in the mouse model (e.g., Saeij et al., Science 2006; Reese et al., PNAS 2011). These effectors are proteins that are secreted from the parasite into the host cell both during (Fig 1) and after (Fig 2) invasion. Understanding the mechanism of action these parasite effectors should yield new insights into the evolution of the host-pathogen relationship on the molecular level, and provide a basis for the development of more effective treatment regimens in human cases of toxoplasmosis.
Approaches: In our lab we exploit the genetic tractability of Toxoplasma to address these questions. We use gene knockouts and knockins to assess the role of key loci in parasite biology. We use microarrays on both host and parasite transcripts to identify genetic networks that are manipulated by the parasite. We use comparative genomics and forward genetics to identify genes that are responsible for phenotypic differences in pathogenesis between strains. We use proteomics to identify previously uncharacterized proteins that Toxoplasma secretes into the host cell. Finally we use well-developed in vivo models to determine the impact of these genes on parasite virulence.
- Sarah Reilly,
- NyJaee Washington, Graduate Student
- Leah Cabo, Graduate Student
- Hisham Alrubaye, Graduate Student
- Rafaela da Silva,
