Dr. Hatfull received his Ph.D. in 1981 with Willie Donachie at the University of Edinburgh, Scotland, performed postdoctoral studies with Nigel Grindley at Yale University and with Fred Sanger at the MRC, and joined the Department in 1988.
https://en.wikipedia.org/wiki/Graham_Hatfull
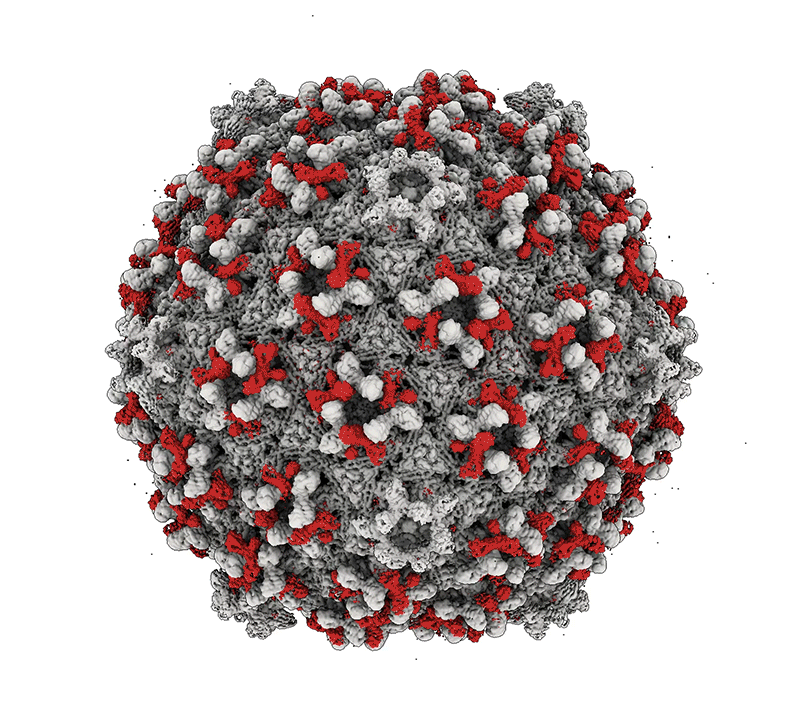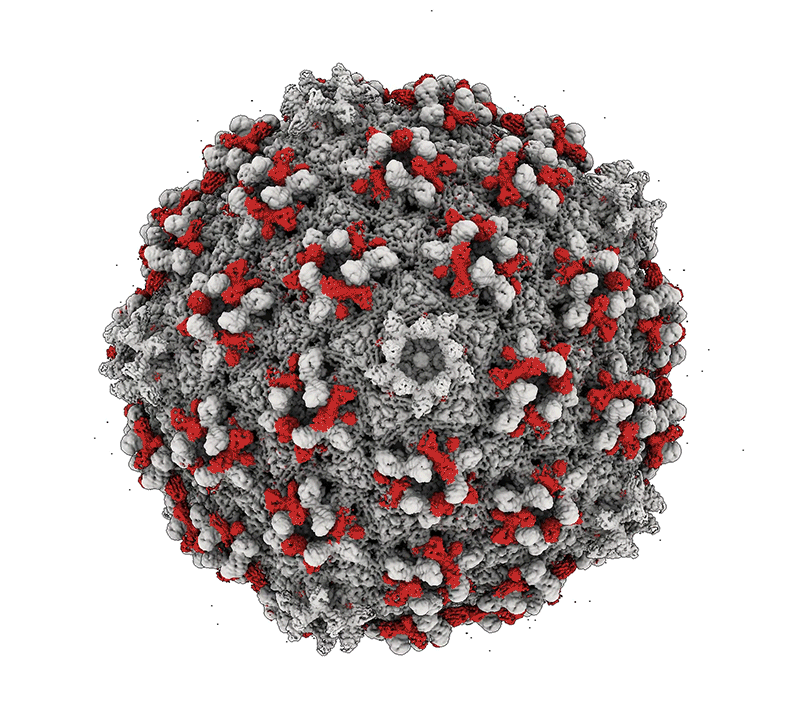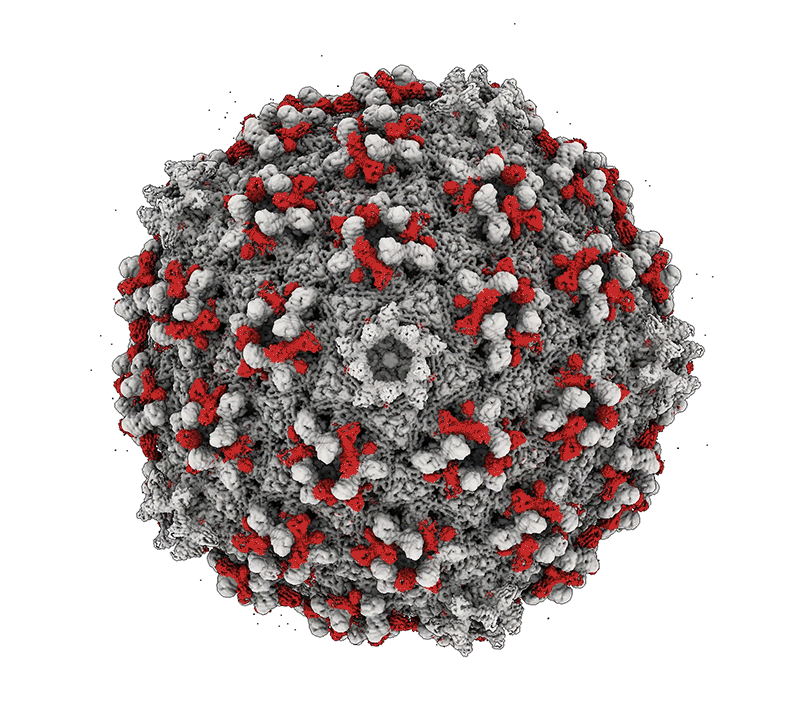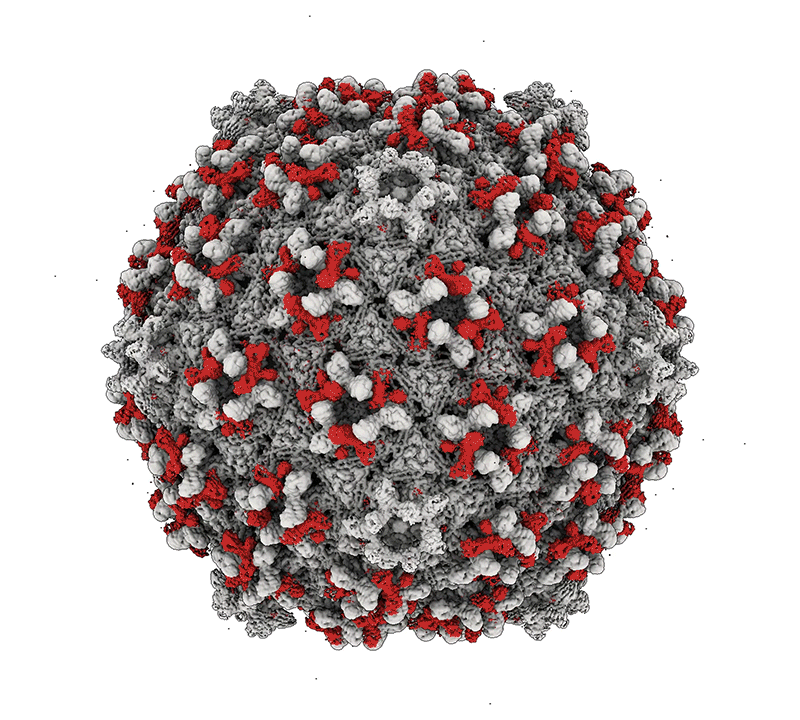
Mycobacteria are among some of the worst pathogens of humankind. Mycobacterium tuberculosis – the causative agent of human TB – infects a substantial portion of the world’s population and is responsible for over 1.5 millions deaths a year. Nontuberculous mycobacteria (NTM) are important opportunistic pathogens and are relatively common in persons with cystic fibrosis and those that are immunosuppressed. Although TB can be treated with multidrug regimens, antibiotic resistance is becoming widespread, and new treatments are needed. NTM infections are intrinsically resistant to many antibiotics, leading to treatment failure and harmful consequences of long-term drug treatments. The study of mycobacteriophages – viruses that infect mycobacteria – can provide new tools for manipulating and understanding the mycobacteria, and the therapeutic use of phages shows considerable promise. Bacteriophage discovery and genomics also offers insights into viral diversity, evolutions, and origins, and provides a powerful platform for science education.

Here's a flavor of some of the current studies going on in the lab:
Bacteriophage genomics. We have been examining bacteriophage genomes since 1988 and through the development of science education programs such as the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program we have established a collection of over 20,000 individual phages isolated on bacteria with the phylum Actinobacteria, half of which were isolated on the strain Mycobacterium smegmatis mc2155. Over 4,000 of the genomes have been completely sequenced and annotated providing a high-resolution view of viral diversity and a wealth of insights into phage biology, diversity, and evolution. A database of these phages and their genomes is available at: https://phagesdb.org.
Understanding mycobacteriophage biology. A curiosity of phage genomes is the high proportion of ‘novel ‘genes of unknown function. We are investigating gene functions using a variety of strategies including genetic studies using gene toxicities, gene expression approaches, and biochemical studies to understand protein activities. We are especially interested in the determinant of phage host range that determine which phages can efficiently infect different bacterial strains. A large and growing collection of clinical isolates of Mycobacterium abscessus is especially valuable for these studies.
Development of tools for genetics. Phage genomes are rich toolboxes for development of new genetic tools. We have developed a suite of integration-proficient plasmid vectors, new selectable markers, and recombineering strategies for use in both fast- and slow-growing mycobacteria. We have also adapted these tools for genetic engineering of the phages, including Bacteriophage Recombineering of Electroporated DNA (BRED) and CRISPY-BRED, which adds a CRISPR-mediated counterselection to the BRED protocol.
Bacteriophage therapy. We are investigating the potential for phages to provide clinical benefits for patients with TB and NTM infections. We have reported the therapeutic use of phages in over 20 patients – mostly with M. abscessus infections – with favorable clinical or microbiological outcomes in a majority of patients. There are many challenges in broadening the potential for phage therapies and advancing from individual compassionate use interventions to clinical trials to understand safety and efficacy.
