Note: Courses make heavy use of laptop and downloaded software. Looking to purchase or borrow a computer? Avoid Chromebooks and choose a laptop with at least 8GB RAM and an i5 processor or equivalent. Install Office 365 (free to Pitt students) before your first class.
Jump to the introductory research lab course list.
"Hands down, best class I took at Pitt!" - Emily Klonicki, Fall 2014/Spring 2015
Introductory research lab courses fulfill requirements of traditional biology labs while students engage in the discovery process of tackling real research. The research questions are generated by faculty, but approaches and methods are feasible for students participating in research for the first time. Every term is a little different from the last; coursework adapts as discoveries are made or challenges are faced in preceding semesters. Courses are designed to maximize student experience and learning while doing meaningful research. Hear from former students and learn more about each course below!
Am I ready to start research?
The simple answer is YES! If you are ready to learn and have an open mind, you are ready for research. Instructors design the course at a realistic level for students and teach the basics in the context of research. Homework assignments will always be meaningful to your research. Will there be hard work? Yes. All biology labs at Pitt are challenging. However, in these courses, the work is towards real research. Some of the courses have prerequisites, so be sure to check that you have completed those as needed.
Real research is for you if...
You want to tackle real-world problems that scientists around the globe are trying to solve.
You want to learn techniques that are used in faculty research labs at Pitt.
You want to make novel discoveries.
How will performing undergraduate research in a lab course help me?
While earning biology lab credit, you'll develop skills and knowledge that are useful, not only in your undergraduate studies, but also in your future endeavors beyond Pitt! You will build critical thinking and scientific reasoning skills as you analyze your data with peers and the course instructor. Additionally, your written and oral communication skills will strengthen as you present your findings to small groups of labmates and at a formal poster session at the end of the course. With guidance, you will choose your own experimental pathway as you learn basic lab skills and continue to grow these talents.
Introductory Research Lab Course Choices
Want to learn more about each lab course? Find the course syllabus here!
Two Semester Course:
|
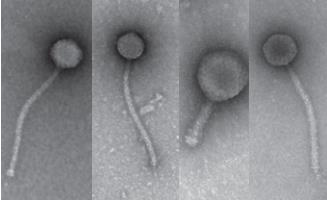
SEA-Phages Virus Hunting Lab Biosc 0058/Biosc 0068 |
 |
Discover and name your own virus |
|
This is a two-semester series for students who plan to take both semesters of introductory biology. In the first semester, you will discover, isolate, characterize and name your own, novel bacteria-infecting virus. You will isolate the DNA from your virus and take pictures of it using an electron microscope. With the rest of your class, you’ll choose some of the viruses for DNA sequencing. In the second semester you learn computer tools to study the information encoded in the DNA and work in teams to generate hypotheses based on the DNA sequence. You’ll also learn to design your own experiments and carry them out to make new discoveries. This course is part of a national research program directed by the laboratory of Dr. Graham Hatfull in the Department of Biological Sciences. Links: SEA-Phage Videos, Graham Hatfull |
||
One Semester Courses:
|
Duckweed Survivor Biosc 0057 |
 |
|
| Western PA is home to a vibrant population of aquatic plants, including species of Duckweed. Duckweed is a known bio-remediator; meaning it has the potential to mitigate the effects of increased concentrations of chemical stressors that are common in local Western PA ponds. Partnering with Martin Turcotte’s lab, you will explore the effects of these chemical stressors on the duckweed population. You will identify the toxicology of pollutants on your duckweed species, analyze competition for resources in a Duckweed community experiment, and use DNA sequence analysis to ID your duckweed plants. Your research will ultimately contribute to our understanding of chemical stressors in the area and how essential aquatic species react to the increasing pollutants. | ||
|
Flower Microbiome Biosc 0057 |
 |
|
| Helianthus, or more well known as the common sunflower, displays a remarkable UV pattern (right). This UV “bullseye” is a known attractor for pollinators and confers select advantages for the flower. While vastly understudied, it is suggested that the UV pattern of Helianthus may also select for certain types of bacteria that grow on the regions of the petals. Partnering with Tia-Lynn Ashman’s lab, you will collect petals from Helianthus sunflowers and grow the petal microbiome. Your job as a scientist will be to isolate one type of bacteria from the petals and discover the identity of your unknown bacteria using DNA sequencing. In addition, you will characterize the bacterial morphology, identify the region of the flower it came from, and determine its sensitivity or resistance to UV light. Ultimately, your research will contribute to our understanding of the flower microbiome, and how UV patterning of flowers may select for certain types of bacterial growth on petals. | ||
|
Water Channels in Disease Biosc 0067 |
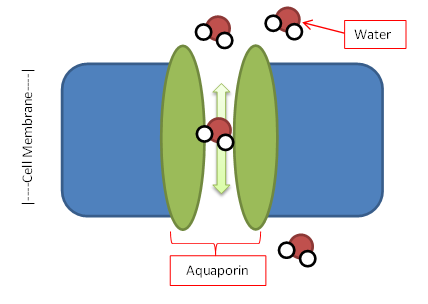 |
Take a look at real gene mutations that cause Diabetes Insipidus |
|
The movement of water in and out of cells is a fundamental cellular process whose disruption can cause disease. In this course you will study mutations in the DNA encoding a kidney membrane water channel called aquaporin 2. People with these mutations have the disease diabetes insipidus, but the reason the mutant aquaporin proteins fail to function is unknown and will be the focus of your research in this course. You will test the production and stability of the protein using our yeast model system. You will also use this model system to develop tests for whether the protein forms a functional water channel and run these experiments to test your own hypotheses of how the mutation may cause disease. This research is connected to work from Dr. Buck and Dr. Kaufmann. Links: Nancy Kaufmann |
||
|
DNA Regulation and Disease |
||
|
Biosc 0067 |
|
Investigate a protein involved in DNA storage that's defective in several human cancers |
|
DNA is tightly and carefully packaged in cell nuclei, but must also be accessible for highly controlled processes such as transcription, replication, and repair. In this course, you will research how DNA packaging and access are controlled and investigate how mis-regulation of a protein involved in these processes can lead to cancer. You will learn to plan, execute, and interpret experiments using common molecular biological techniques as you generate novel mutant proteins that affect DNA organization using yeast as a model organism. You will also read current scientific literature to understand how the proteins involved in DNA packaging result in human disease and consider how your findings could apply to human disease studies. Links: Karen Arndt |
||
|
Neurobiology: Cell Morphogenesis Biosc 0067 |
|
Discover genes that control cell shape |
|
During human fetal development, a flat layer of cells bends to form the neural tube. Failure to properly form this tube leads to severe brain and spinal cord defects. In this course, you will research a protein called Shroom which regulates cell shape changes in most animals. Mice without the Shroom3 protein have neural tubes which “mushroom out” during development and fruit flies with extra Shroom protein have defective eyes and wings. You will search for proteins that cooperate with Shroom to change cell shape in fruit flies using genetic approaches and assessing for changes in eyes and wings. You will develop hypotheses about which proteins work with Shroom and how they might control shape change and use microscopy to test your predictions. This work is directly connected to research in Dr. Jeff Hildebrand’s lab. Links: Jeffrey Hildebrand |
||