
Contact
A critical question to ask, particularly in this genomic era, is how organisms interpret the vast amounts of information encoded in their genomes. The Arndt lab studies the first step in gene expression, the synthesis of mRNA by RNA polymerase II, with a focus on the mechanisms that regulate transcription in the chromatin environment of a eukaryotic cell. The fundamental importance of understanding transcriptional regulation is evident from the large number of human developmental defects and diseases, including cancer and AIDS, that arise when cellular transcription factors are altered by mutation or commandeered by viral proteins.
The transcription cycle can be divided into three steps: (1) promoter recognition and initiation of RNA synthesis; (2) elongation of the growing RNA chain; and (3) termination. Our current studies focus on the elongation and termination steps in the cycle. In our research, we exploit a vast array of genetic, biochemical, and genomic tools uniquely available in the model eukaryote, Saccharomyces cerevisiae. This simple yeast is an excellent model system for our studies because the proteins and mechanisms that regulate transcription have been highly conserved from yeast to man.
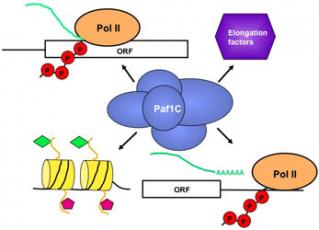
During transcription elongation, RNA polymerase II encounters obstacles, notably nucleosomes, which block its path. Eukaryotes express proteins that facilitate or impede polymerase movement by altering the chromatin template. One of these proteins is the conserved Paf1 complex. The Paf1 complex associates with RNA polymerase II on the coding regions of transcribed genes, interacts with other highly conserved elongation factors, couples histone modification to transcription elongation, and controls the efficiency of transcription termination. Thus, the Paf1 complex is a multifunctional regulator of gene expression that lies at the intersection between transcription elongation and chromatin modification. Current projects in the lab are focused on:
- The mechanisms by which the Paf1 complex regulates transcription elongation and events coupled to this process, especially RNA 3' end formation
- The genes that are regulated by the Paf1 complex and how the Paf1 complex is recruited to these genes
- The mechanisms by which the Rtf1 subunit of the complex controls histone modifications
- The functions of the Cdc73 subunit of the Paf1 complex, a protein with tumor suppression activity in human cells
- The cellular factors, including a novel ubiquitin protein-ligase, that protect the cell against the consequences of deregulated transcription
- Margaret Shirra, Postdoctoral Researcher
- Alex Francette, Graduate Student
- Sarah Tripplehorn, Graduate Student
- Sanchirmaa Namjilsuren, Graduate Student
- Tasniem Fetian, Graduate Student
- Aakash Grover, Graduate Student
