
Contact
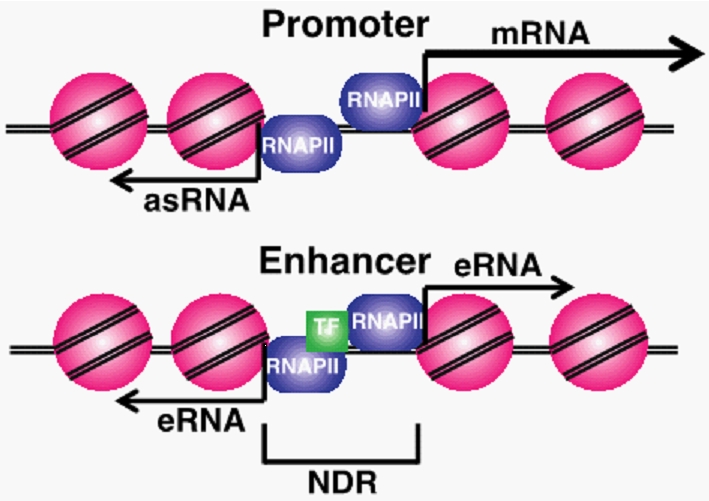 |
Figure 1. Diagram of promoter and enhancer regulatory regions. Promoter proximal antisense RNA (asRNA), coding gene (mRNA), enhancer RNAs (eRNAs) and nucleosome depleted regions (NDR) are labeled. Pink circles are nucleosomes and purple circles are RNAPII. |
An unexpected finding from genome-scale studies is that the majority of the human genome is transcribed. Although protein-coding regions comprise only ~2% of the human genome, at least 75% is transcribed at detectable levels. These findings have led to a re-evaluation of the mammalian genome – if non-coding regions are transcribed, the resulting non-coding RNAs (ncRNAs) may have important functions. This possibility has tremendous ramifications for biomedical research, since clinical samples subjected to diagnostic sequencing are typically examined at only a subset of important genes, and only in their coding sequences.
ncRNAs are produced from many different regulatory regions in cells (Fig 1). Short ncRNA molecules expressed from within enhancer sequences, termed eRNAs, are thought to be necessary for enhancer looping and gene regulation. Promoter associated ncRNAs originating upstream from promoters (PROMPTs) have been identified in numerous eukaryotes, and mechanisms of their termination and degradation have been described. However, the transcriptional regulation and function of the majority of these ncRNA transcripts remain largely undefined.
One key regulatory mechanism shared among eukaryotes is the control of access to regulatory sequences by transcription factors through alteration of nucleosome occupancy or positioning. Nucleosome remodeling factors use the energy from ATP hydrolysis to reposition, deposit, or remove nucleosomes at regulatory regions by altering histone-DNA contacts. The actions of nucleosome remodeling factors are critical for transcription, DNA repair, and other essential cellular functions. Given their key roles in regulation of gene expression and genome integrity, it is perhaps not surprising that nucleosome remodeling factors are among the most commonly mutated or epigenetically silenced genes in human cancers and neurological disorders. However, the mechanisms by which loss of nucleosome remodeling factors function contributes to cancer and disease development are largely unknown.
Recently, we found that the embryonic stem (ES) cell-specific nucleosome remodeling complex esBAF, which occupies both enhancers and promoters, is required for regulating ncRNA expression throughout the ES cell genome (Fig 2). ES cells must carefully regulate the decision to either self-renew (proliferate as ES cells) or differentiate into precursors of the 200+ cell types found in adult humans and at the heart of this decision is the ES cell gene regulatory network, which is regulated by the coordinated efforts of numerous proteins including chromatin regulatory complexes such as esBAF. Determining the mechanism factors utilize to regulate the self-renewal- or differentiation-specific gene network is essential to uncover basic developmental processes and further our understanding of ES cell-based therapeutics.
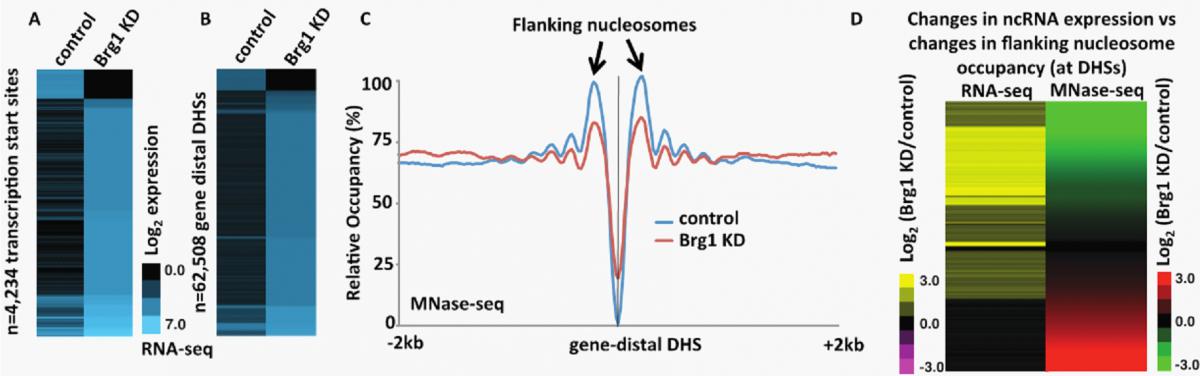 |
| Figure 2. esBAF represses ncRNA expression by promoting nucleosome occupancy flanking NDRs. (A-B) Heatmap of strand-specific RNA-Seq showing transcripts generated antisense to TSSs (A) or surrounding gene distal DHSs (B) in control and Brg1 KD ES cells. These data show a large increase in the expression of transcripts generated in Brg1 KD cells relative to control. (C) Aggregation plot of MNase-Seq data showing nucleosome occupancy at gene distal DHSs in control and Brg1 KD ES cells. These data show a decrease in NDR-flanking nucleosome occupancy in Brg1 KD cells. (D) Fold change of Brg1 KD/control gene distal transcripts identified through RNA-Seq (left panel) and flanking nucleosome occupancy identified through MNase-Seq (right panel) paired and sorted. These data show that decreased flanking nucleosome occupancy (green) correlates with increased transcript production (yellow) in Brg1 KD cells. |
The Hainer lab will address a number outstanding questions regarding transcription regulation in ES cells, utilizing a variety of molecular, cytological, and genomic techniques, through distinct but related projects. Specifically, we will determine (1) the network of nucleosome remodeling complexes regulating ncRNA expression, (2) how esBAF and other nucleosome remodeling complexes regulate higher order chromatin structure, (3) the functions of enhancer-specific ncRNAs in enhancer looping, gene regulation, and control of the ES cell state, and (4) the mechanisms underlying regulation of mRNAs by enhancer and promoter ncRNAs.
- Benjamin Patty, Graduate Student
- Sarah Tripplehorn, Graduate Student
- Santana Lardo, Research Specialist III
- Rithika Sankar, Graduate Student
- Mika Wesley, Undergraduate Researcher
- Aigbirhemwen Woghiren-Afegbua, Graduate Student
- Emmy Brown, Graduate Student
- Kya Foxx, Undergraduate Researcher
- Bryona Jackson, Undergraduate Researcher
<p><strong>DC Klein, SM Lardo, SJ Hainer. </strong
DC Klein, SM Lardo, KN McCanne
DC Klein, K Troy, SA T
H Zou, B Poore, EE Brown, J Qi
C McCann, M Quinteros, I Adelugba, M Morgada, A
SM Lardo and SJ Hainer
BJ Patty and SJ Hainer.
BJ Patty and SJ Hainer. Non-Coding RNAs and Nuc
K Troy, Y. Liu*, SJ Hainer*. PathSTORM: a road
SJ Hainer* and CD Kaplan* Specialized RSC: subs
J Xu, H Ma, H Ma, W Jiang, M Duan, S Zhao, C Ga
NA Fraunhoffer, N Agarwal, K Takeishi, A Ostrow
DC Klein and SJ Hainer. Chromatin Regulation an
DC Klein and SJ Hainer. Genomic Methods in Prof
C Tavera-Montanez*, SJ Hainer*, D Cangussu, SJV
SJ Hainer, A
SJ Hainer and TG Fazzio. High‐Resolution C
D Acharya, SJ Hainer
SJ Hainer, KN M
SJ Hainer and JA Martens. Regu
SJ Hainer and TG Fazzio. Regul
SJ Hainer, W Gu, BR Carone, BL
PB Chen, LJ Zhu, SJ Hainer, KN
BR Carone, JH Hung, SJ Hainer,
SJ Hainer, BA Charsar, SB Cohe
JA Pruneski, SJ Hainer, KO Pet
SJ Hainer and JA Martens. Iden
SJ Hainer and JA Martens. Tran
SJ Hainer, JA Pruneski, RD Mit
2021 Iris Marion Young Award, University of Pittsburgh
2020 “Most Valuable Professor”, Awarded by University of Pittsburgh Women’s Diving Team
2020 University nominee for Pew Biomedical Scholar
2019 University nominee for Edward Mallinckrodt, Jr Foundation Scholar Program
2019 University nominee for Johnson&Johnson WiSTEM2D Scholars Award
2019 University nominee for Packard Fellowships for Science and Engineering
2018 Nominee for Arnold and Mabel Beckman Foundation Young Investigator Award
2016 - 2019 Special Fellow, Leukemia and Lymphoma Society
2013 - 2016 Fellow, Leukemia and Lymphoma Society
2012 - 2013 T32 Institutional Training Grant Postdoctoral Fellowship
